

Alys Clark (University of Auckland), Sara Loo (Johns Hopkins University), Fiona R. Macfarlane (University of St Andrews), and Thomas Woolley (Cardiff University).
- In Memory: Torcom Chorbajian, Long-time Volunteer and SMB Officer
-
News – updates from:
- People – An interview with Professor Jae Kyoung Kim, who will be a plenary speaker at the Joint Annual Meeting of the Korean Society for Mathematical Biology and Society for Mathematical Biology in Seoul this year.
- Editorial – on 'A look forward to KSMB - a chat with Dr Yangjin Kim' on the upcoming SMB-KSMB conference.
- Featured Figure – Highlighting the research by early career researcher Ryan Murphy, University of Melbourne.
To see the articles in this issue, click the links at the above items.
Contributing Content
Issues of the newsletter are released four times per year in Spring, Summer, Autumn, and Winter. The newsletter serves the SMB community with news and updates, so please share it with your colleagues and contribute content to future issues.
If you have any suggestions for content or on how to improve the newsletter, please contact us at any time. We appreciate and welcome feedback and ideas from the community. The editors can be reached at newsletter@smb.org.
We hope you enjoy this issue of the newsletter!
Alys, Sara, Fiona, and Thomas
In Memory: Torcom Chorbajian: Long-time Volunteer and SMB Officer
Contributed by Lou Gross, Ray Mejia and John Jungck
The Society lost a tremendous long-standing leader and volunteer on January 19, 2024 when Torcom Chorbajian passed away in Lafayette, Colorado. Without any paid staff members, except for those associated with the Bulletin, SMB has functioned over its history due to the dedicated efforts of many volunteers. Torcom was exemplary as the volunteer Treasurer and Board member for nearly forty years. He had tremendous knowledge of the history of the Society and had direct personal connections with the leadership over the first decades, by far attending more Board meetings than anyone else in our history.
As the person who managed membership records, dealt with all financial aspects of the Society and handled interactions with the various publishers of the Bulletin, Torcom devoted untold hours to Society business. He was the one who corresponded with our far-flung membership for decades, handling all kinds of membership and subscription challenges. He took personal interest in our members, particularly those outside North America who relied on their subscriptions to stay informed of the latest research in the time before electronic connections were prevalent. Many were the times he called the current Society President to discuss challenges faced by one of our members and to express concern about sometimes delayed responses from the various publishers. He knew our membership far better than anyone else in the leadership and had attended all of our Annual Meetings for many years. For many meetings, he would manage the display of collections of books to provide our members an opportunity to see the latest texts from various publishers. He handled the distribution of travel funds for many, many students over the years who benefited from the Society’s Landahl awards to enable them to attend meetings (see the photo attached from the 2010 Rio meeting of Torcom writing a check for a student attendee). Torcom also designed the three-sided pens (see photo) handed out at many meetings that are still cherished by members due to the way they sit so comfortably in your hand.


Torcom’s contributions were celebrated in 2008 with the first Torcom Chorbajian Lecture at the Annual Meeting at the University of Toronto. As then-President Gerda de Vries noted in the January 2013 SMB Newsletter “I want to thank Torcom Chorbajian for serving as Treasurer for almost 40 years. Torcom has been a member of the SMB since its inception in the 1970s and was appointed Treasurer two years later. Torcom has accepted the title of Honorary Treasurer of the SMB.” Tocom is remembered as a friend and tireless steward of the SMB and its members.
News Section
By Fiona Macfarlane

SMB Subgroups Update
Cell and Developmental Biology Subgroup
The Cell and Developmental Biology (CDEV) subgroup was active (with minisymposia, contributed talks, posters, a subgroup business meeting, and a group dinner) at the 2023 SMB Annual Meeting in Columbus, OH, and we're looking forward to the 2024 SMB Annual Meeting in Seoul, Korea!
In addition to our activities at the annual meetings and our blog-post series (https://smb-celldevbio.github.io/blog/) highlighting researchers in our community, we started two new virtual initiatives in the last year. First, we held mentored mock virtual interviews for students and postdocs preparing for the academic job market (thanks to all who participated as mentors and mentees!). Second, we are holding our first virtual micro-conference “Virtual Cell and Development Festival Week” from March 18–21, 2024. The schedule features plenary talks on research and professional development topics, several minisymposia, and two panels (focused on industry careers and the future of models and software platforms). Each day of the festival week has about 2 hours of programming, with a range of times selected to fit many timezones. Please see https://smb-celldevbio.github.io/cdevfestival/ for more information and registration details (registration is free). All are welcome and encouraged to attend our first virtual CDEV festival week!
Immunobiology and Infection Subgroup
The Immunobiology and Infection subgroup is excited to host four outstanding speakers in its annual minisymposium at the joint Annual Meeting of the KSMB and SMB, in addition to the many excellent sessions being organized by our members. Join us for talks by Reginald McGee, Wasiur Khuda Bukhsh, Adrianne Jenner, and Past Chair Stanca Ciupe. Hope to see you in Seoul!
At last year’s SMB Annual Meeting at Ohio State University, together with SMB and the National Institute of Allergy and Infectious Diseases (NIAID), our subgroup co-organized a half-day workshop Bridging multiscale modeling and practical clinical applications in infectious diseases. This event brought together top experts in multiscale mathematical modeling with experimentalists and clinicians working at the frontier of immunity and infectious diseases to share their research and discuss challenges and opportunities for future work. We were thrilled to see the high level of interest from conference attendees and are looking forward to organizing a future iteration of the event. If you missed it, or want to relive the fun, the organizing team wrote a summary article which will be forthcoming in the Bulletin of Mathematical Biology, keep an eye out and we will send around when it is published. Thank you to the co-organizers, speakers, and participants.
Mathematical Epidemiology and Mathematical Oncology Subgroups
The Mathematical Epidemiology (MEPI) and Mathematical Oncology (ONCO) subgroups hosted SMB MathEpiOnco 2024, a joint virtual mini-conference February 18-20. Over 150 registered participants from 22 countries attended the three-day meeting. The conference featured plenary talks by Marisa Eisenberg (University of Michigan, USA), Claudia Pio Ferreira (São Paulo State University, Brazil), and Natalia Komarova (U.C. San Diego, USA) as well as a panel discussion of Opportunities at the Interface of Mathematical Epidemiology and Oncology with panelists Hanna Dueck (National Institutes of Health, USA), Zhilan Feng (National Science Foundation and Purdue University, USA), and Ami Radunskaya (Pomona College, USA) and a tutorial session on Stochastic Processes in Epidemiology and Oncology led by Linh Huynh (Dartmouth College, USA) and Pujan Shrestha (Texas A&M, USA). In addition, the conference featured 22 contributed talks for SMB members working on problems in mathematical epidemiology, mathematical oncology, and the intersection of the two fields.

The conference prompted important discussions about similarities in approaches to studying problems in oncology and epidemiology with a mathematical lens and highlighted areas for research growth in questions that are relevant to both fields. Furthermore, the conference underscored important links between infectious disease and cancer that leave open a number of interesting questions that mathematical biologists can explore. The conference closed with a period of discussion in working groups with goals such as Connecting within-host dynamics with population level incidence and transmission dynamics, Investigating the role of models in studying infectious diseases that lead to cancer, and Connecting models and parameters across different types of model structures. Participants in working groups made plans for future projects and mini-symposia to be organized for future scientific meetings.
SMB MathEpiOnco 2024 was organized by Jason George (Texas A&M), Meredith Greer (Bates College, USA), Linh Huynh (Dartmouth College), Harsh Jain (University of Minnesota Duluth, USA), and Michael Robert (Virginia Tech, USA). For more information, visit the conference website:https://seminar.math.vt.edu/SMB-MEPI-ONCO/schedule.html
Upcoming Conferences and Workshops
Society for Mathematical Biology Annual Meeting
From 30th June - 5th July Friday 2024, the joint annual meeting of the Korean Society for Mathematical Biology and the Society for Mathematical Biology will be held at KonKuk University, Seoul, Republic of Korea. Early bird registration will be open until 30th April 2024, for more details check the conference website: https://smb2024.org/
Royal Society Publishing
.jpg)
.tif) The following Royal Society theme issue has been highly cited, downloaded and is FREE to access online: ‘Technical challenges of modelling real-life epidemics and examples of overcoming these’ compiled and edited by Dr Jasmina Panovska-Griffiths, Dr William Waites, and Professor Graeme J Ackland - see https://royalsocietypublishing.org/toc/rsta/2022/380/2233
The following Royal Society theme issue has been highly cited, downloaded and is FREE to access online: ‘Technical challenges of modelling real-life epidemics and examples of overcoming these’ compiled and edited by Dr Jasmina Panovska-Griffiths, Dr William Waites, and Professor Graeme J Ackland - see https://royalsocietypublishing.org/toc/rsta/2022/380/2233
.jpg)
The COVID-19 pandemic has highlighted the importance of mathematical modelling in informing and advising policy decision making. Via a collection of sixteen papers, this issue showcases how the Royal Society coordinated efforts of diverse scientists to help model the coronavirus epidemic and overcome a number of technical challenges. Different papers address the utilisations of different technical modelling frameworks and how different techniques are combined, show how modelling of different scenarios can give informed scientific advice, discuss how to correctly quantify the uncertainty of the model parameters and projections, and flag up the importance of transparency and robustness of models and numerical code to ensure reproducibility of the results. Read more in a blog post by one of the Guest Editors: https://royalsociety.org/blog/2022/08/modelling-epidemics-ramp/
We are also looking for new theme issues and that if you are interested in submitting a proposal, please visit the website https://royalsocietypublishing.org/rsta/guest-editors or contact the Editorial Office for more information - philtransa@royalsociety.org.
Back to the top
People
By Alys Clark

We interviewed Professor Jae Kyoung Kim, Chief Investigator of the Biomedical Mathematics Group at the Institute for Basic Science, Republic of Korea and an academic staff member in Mathematics at the National University in Daejeon, Republic of Korea, find out more here.
Back to the top
Editorial

By Sara Loo
A look forward to KSMB – a chat with Dr Yangjin Kim
As the year continues to tick quickly on and winter comes to an end, we can look forward to many exciting things. Not least of these is the upcoming Korean Society of Mathematical Biology – Society of Mathematical Biology joint meeting in July. In the lead up to this summer’s conference, and as submissions for minisymposia and contributed talks come streaming in, I met with Yangjin Kim, co-chair of the meeting’s organising committee to get to know a bit more about KSMB and what we can expect in Seoul in July.
On a wintery evening over Zoom, we chatted about the history of KSMB. Founded in 2005, its co-founders quickly established bridges across disciplines – one of the founders, Tae-Soo Chon is a biologist. This quickly and firmly founded the society within the biological science community, as well as in its natural habitat in the mathematical sciences. This led to many natural and fruitful collaborations. Starting from very small numbers, the society now draws in over 150 participants at their annual meetings. Beyond this, the society has been a strong advocate for the field in the Asia region, convening the China-India-Japan-Korea Conference on Mathematical and Theoretical Biology last year in Jeju Island, South Korea.
Having been a part of SMB since early in his research career, Yangjin speaks fondly of his experiences at SMB conferences. His passion for creating a similar environment for others across many different regions is evident. “I grew up with SMB”, he mentions. He has only missed two meetings since his first in Raleigh in 2005, and as a PhD student he earned himself a Landahl Travel Grant in 2006. These influences are long-reaching, and throughout our conversation he is reminded of how beneficial the society has been to him – “I always feel comfortable when I attend the SMB meeting every year… having a chance to talk to people in my research area and getting [to meet] mentors.” He has encouraged his students to attend the yearly meetings and “it has been wonderful for them, they say.”
He tells me how fruitful his interactions with SMB members have been. He has worked with many mentors and peers throughout his career and time as an SMB member – his thesis advisor Hans Othmer, Avner Friedman, Mark Chaplain, amongst others. Being a part of a community of like-minded peers and receiving advice, feedback and, even, criticisms from others in the society have marked his career. In some sense, he “[sees] it as the center of [his] career.”
This sort of environment is something he values greatly, and something he seeks to share with researchers all over the world. I ask him what he hopes the conference will be like, or something he hopes it may achieve. “I want it to be really international”, he says. Though Yangjin trained in the US and spent 13 years there in his early career, his position at Konkuk University in South Korea has allowed him to grow and cultivate excitement and interest for the field in Asia. Throughout Asia, other regional societies of mathematical biology have popped up, and are starting to grow – the Phillipine Society for Mathematical Biology was launched earlier this year, and the CIJK conference last year was its eighth iteration. Holding KSMB-SMB in Seoul will be a great foundation for these smaller societies to gain support and interact with members of our larger community, stimulating ongoing research in the region. Already, they have seen a range of diverse abstracts from many Asian countries and young scientists.
So what else can we look forward to in Seoul in July? Yangjin mentions excellent food, beautiful modern and traditional buildings, some K-pop, a beautiful campus and perhaps some drone flying is on the cards. Beyond that, take a moment to be thankful for the joint meeting and the commitment of the organising committee in pulling this off. This was originally planned for 2022 and though it was inevitably postponed, here we are 2 years later. “It is a long time coming, [we] almost got exhausted from asking ‘when are we going to have this!’”, Yangjin laughs. It took many years of preparation, discussions have seen multiple SMB presidents, and ongoing commitment from the SMB board and members of KSMB. 감사합니다 See you in July!
Back to the top
Featured Figure
By Thomas Woolley
Early Career Feature - Ryan Murphy, University of Melbourne
In this issue, we feature the article “Formation and growth of co-culture spheroids: New compartment-based mathematical models and experiments”. This research was performed by Ryan J. Murphy (University of Melbourne), Gency Gunasingh (University of Queensland), Nikolas K. Haass (University of Queensland), and Matthew J. Simpson (Queensland University of Technology).
Tumour spheroid experiments are routinely performed to investigate cancer progression and test anti-cancer therapies. In our previous studies, we have connected the seminal Greenspan mathematical model to monoculture tumour spheroid growth data for the first time, leading to practical experimental design recommendations and quantification of the time evolution of spheroid structure (necrotic core, proliferation-inhibited intermediate region, proliferating rim). By considering time-dependent oxygen conditions, we also revealed that tumour spheroids can experience surprising necrotic core dynamics and transient reversal of growth phases that had been well-characterised for over fifty years.
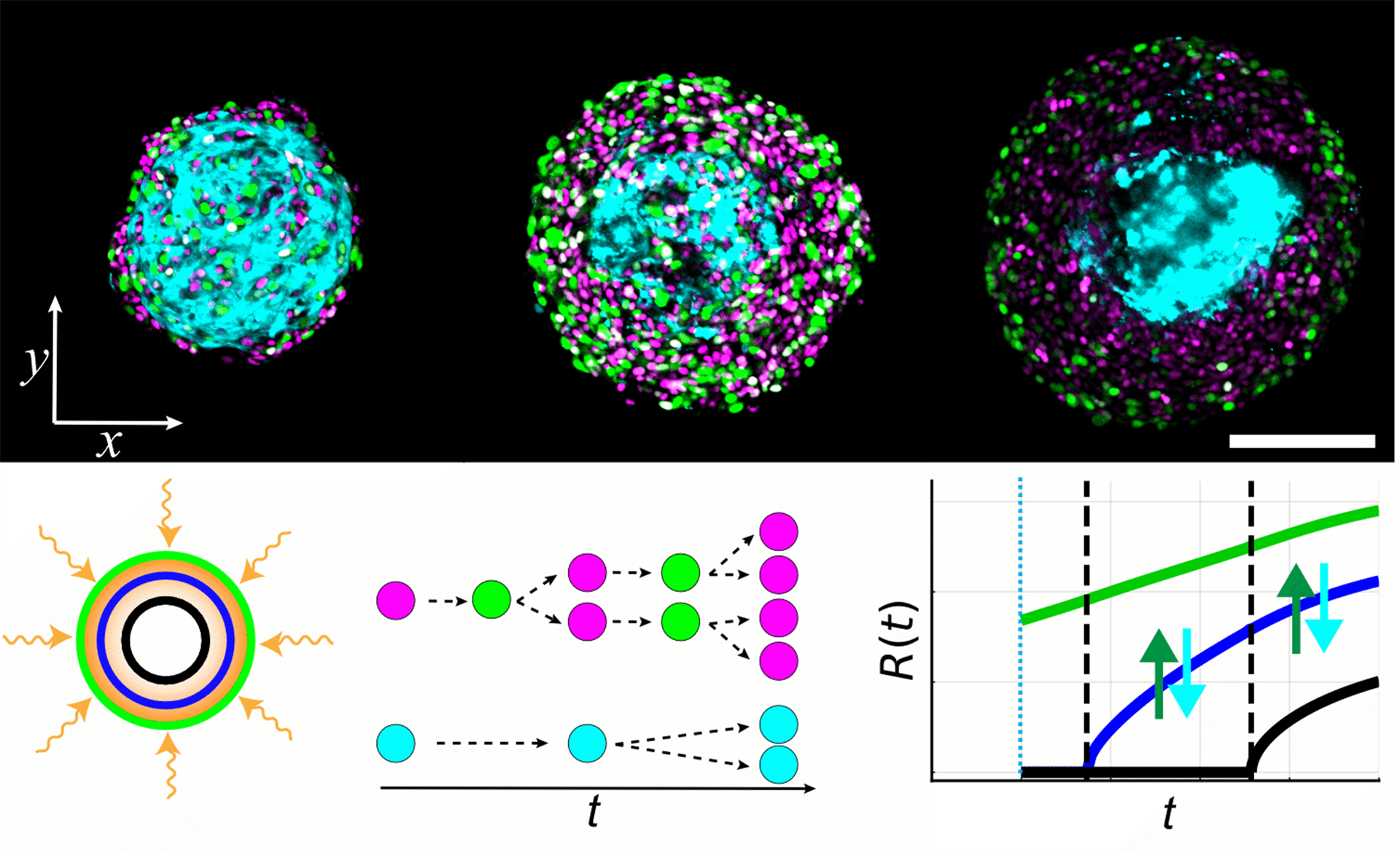
In this study, we consider co-culture tumour spheroid growth experiments. Co-culture spheroid experiments are challenging to interpret as they are comprised of two or more cell types that may have different characteristics, such as differing proliferation rates or responses to nutrient availability. The dynamics are further complicated by multiple biological processes occurring on overlapping timescales. As Greenspan’s model has been valuable in analysing monoculture spheroid data, we first connect Greenspan’s model to co-culture data for the first time. We find that parameter estimates are consistent for co-culture spheroids seeded with different initial proportions of two cell types. However, since the model assumes all cells behave identically, it cannot capture experimentally observed internal dynamics of growing co-culture spheroids.
For greater insights, we generalise a class of compartment-based mathematical models previously restricted to spheroids composed of one cell type, such as Greenspan’s model, so that they can be applied to spheroids consisting of multiple cell types. It is then straightforward to develop and explore multiple natural two-population extensions to Greenspan’s seminal model, where the populations may differ with respect to their proliferation rate, death rate, response to nutrients, or migration preferences. By connecting these new models to data, we reveal biological mechanisms that can describe the internal dynamics of growing co-culture spheroids and those that cannot. This mathematical and statistical modelling-based framework is well suited to analyse spheroids grown with multiple cell types, and the new class of ordinary differential equation-based mathematical models provide opportunities for further mathematical and biological insights.
You can find out more about this research here: https://link.springer.com/article/10.1007/s11538-023-01229-1
Back to the top
